January 14, 2015
by Rob Mitchum, Computation Institute
Hydrated excess protons, also known as H3O+ and hydronium cations, are skilled escape artists. In a space full of water, the extra protons can hop from H2O to H2O like an action star leaping train cars. If they find a hydrated channel, protons can use that water-hopping skill to shoot across membranes, as well. Now, new computational chemistry research from the laboratory of Gregory Voth, professor of chemistry, finds that protons don’t even need those tunnels to be filled with water first--they can actually create their own “water wire” to span the channel and enable their thrilling escape.
The study, which appeared last month in the Journal of Physical Chemistry, adds a new wrinkle to the 208-year-old theory that first predicted this proton-hopping behavior. In 1806, German chemist Theodor Grotthuss predicted that protons would move from water molecule to water molecule in a sort of “bucket line,” where each water would give up a proton at the same time as it receives a new one. Even though Grotthuss made this prediction when water was incorrectly thought to be made up of just one hydrogen and one oxygen, modern experimental evidence has upheld its general principles.
This unique travel mechanism makes protons--also known as “positive charge defects”--atomic speed demons relative to other ions, said co-author Jessica Swanson, senior research scientist at the University of Chicago. “The positive charge defect can move very long distances without any nucleus moving more than a quarter angstrom,” Swanson said. “That enables an excess proton to diffuse through water much more quickly than any other ions.”
But this clever strategy supposedly screeches to a halt when a proton reaches a dry, hydrophobic channel through a membrane--with no water to hop between, the proton seems to be trapped. Because these hydrophobic channels are common in many biological proteins, chemists have used computational models to try to figure out how protons overcome this obstacle, usually concluding that they have to wait until water conveniently and spontaneously fills the channel, forming a “water wire” to smooth the proton’s passage.
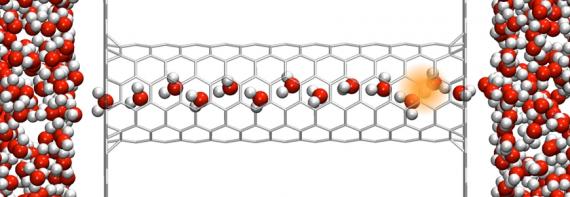
A team from the Voth-Swanson collaboration enlisted Yuxing Peng (then a graduate student in chemsitry, and now a scientific computing consultant with the Research Computing Center) to look at the energy profile as the proton passes through a channel in such circumstances. Peng used a molecular dynamics model on Midway, the University's supercomputing cluster, to create a virtual system where a sheet of graphene separates two water-filled spaces, with only a hydrophobic carbon nanotube connecting the two sides. (The collaboration made use of the RCC's Cluster Partnership Program, which provided them with the dedicated computational resources they needed; this research alone consumed almost 1 million service units, equivalent to 100 processors running continuously for a month and a half.) They then began modeling the motion of a proton and the energy barriers it faces in this system--when they discovered a surprise.
“We found this interesting phenomenon,” Swanson said. “When the proton reached the mouth of the tube, water started shooting through it, filling the tube until it was completely wet. Then, of course, it was much easier for the proton to go through.”
Essentially, the protons were creating their own water wire, paving the road ahead of them so they could get to the other side. Intriguingly, the researchers discovered that they did this through a previously unobserved twist on the Grotthuss mechanism. Instead of a proton hopping between water molecules, it does the opposite, sitting at the opening of the channel and hopping without moving, to propel waters ahead of it. Once the chain of water molecules reaches the other side (or at least a protonatable residue on an amino acid inside the channel), the proton has established its path of escape.

The new mechanism offers a possible explanation for the abundance of hydrophobic channels in biological proteins that are known to transport protons, such as the proton-chloride antiporter and cytochrome c oxidase, both found in mitochondrial and bacterial membranes. It also serves as a technical reminder that you can’t cut certain corners in computational models of molecular activity, as an incomplete system would have missed this new phenomenon.
“We’re seeing in a lot of systems you really have to sample both the water solvation and the charge migration at the same time or you get skewed results,” Swanson said. “The implication of course, is that we need to stop thinking that water wires must exist, preformed, before one can even consider proton transport.”
See the original article at https://www.ci.uchicago.edu/blog/how-hydrated-excess-protons-make-their-escape.